I did write about an interesting paper last year concerning calcium channels and intellectual disability; it was from a city in China called Changsha.
Epiphany: Calcium channelopathies and
intellectual disability
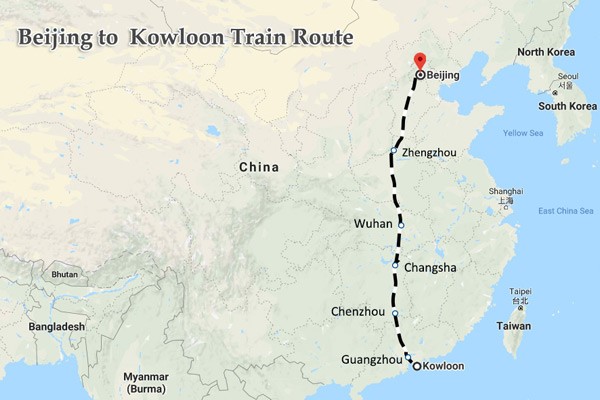
Changsha is on the old
train line and the new high speed line from Beijing to Hong Kong. So like many
other people, I must have passed by this city of 10 million on the old line, as
a backpacking student many years ago.
After three years of
closure, China announced that it is reopening to foreign visitors. China is
well worth a visit and their high speed trains make travel much easier than it
used to be.
Before moving on to today’s paper, I will mention the case study below from one of China’s top hospitals, the PLA hospital in Beijing. They used the well known mTOR inhibitor Rapamycin to successfully treat an 8 year old boy with idiopathic (of unknown cause) autism. This drug has been used in models of autism. The mTOR inhibitor Everolimus is approved as adjunctive therapy for a single gene autism called TSC to treat seizures. Click on the link below to read the one page case report.
Some readers have mentioned this case study and at least one has made a trial. In that case the drug was well tolerated but did not moderate autism symptoms.
Mammalian target of
rapamycin (mTOR) regulates cell proliferation, autophagy, and apoptosis by
participating in multiple signaling pathways in the body. Studies have shown
that the mTOR signaling pathway is also associated with cancer, arthritis,
insulin resistance, osteoporosis, and other diseases including some autism.
Today we return to
Changsha for another interesting paper about the altered immune system in
autism and other neurological conditions.
It is an interesting study because it is based on samples from 2,500 brains of controls and patients with six major
brain disorders - schizophrenia, bipolar disorder, autism spectrum disorder,
major depressive disorder, Alzheimer’s disease, and Parkinson’s disease.
One of
the reasons so little progress has been made in treating any neurological
condition is the inability to take physical samples to experiment with. All the 2,500 brain samples are taken from brain
banks, not live people.
When it comes to autism that means the sample likely reflects severe autism (DSM3 autism). No self-identified autism in today’s samples, their brains are unlikely to be donated to medical science.
Immunity-linked genes expressed differently in brains of autistic people
Genes involved in immune system function have atypical
expression patterns in the brains of people with some neurological and
psychiatric conditions, including autism, according to a new study of thousands of
postmortem brain samples.
Of the 1,275 immune genes studied, 765 — 60 percent —
showed elevated or reduced expression in the brains of adults with one of six
conditions: autism, schizophrenia, bipolar disorder, depression, Alzheimer’s
disease or Parkinson’s disease. The expression patterns varied by condition,
suggesting that there are distinct “signatures” for each one, says lead
researcher Chunyu Liu, professor of psychiatry and behavioral sciences at
Upstate Medical University in Syracuse, New York.
The expression of immune genes could potentially serve
as a marker for inflammation, Liu says. Such immune activation — particularly
while in utero — has been associated with autism, though the mechanisms are far
from clear.
“My impression
is the immune system is not really a very minor player in brain disorders,” Liu
says. “It is a major player.”
It’s impossible to discern from this study whether
immune activation played a role in contributing to any condition or whether the
condition itself led to altered immune activation, says Christopher
Coe, professor emeritus of biopsychology at the
University of Wisconsin-Madison, who was not involved in the work.
“A study of the postmortem brain is informative,” Coe
says. “But not definitive.”
Liu and his team analyzed the expression levels of 1,275 immune
genes in 2,467 postmortem brain samples, including 103 from autistic people and
1,178 from controls. The data came from two transcriptomics databases — ArrayExpress and the Gene Expression Omnibus — and other
previously published studies.
Brains from
autistic people had, on average, 275 genes with expression levels that differed
from those of controls; brains from people with Alzheimer’s disease had 638 differentially
expressed genes, followed by those with schizophrenia (220), Parkinson’s (97),
bipolar disorder (58) and depression (27).
Autistic men’s
expression levels varied more than those of autistic women, whereas the
brains of women with depression showed more variation than those of men with
depression. The other four conditions showed no sex differences.
The
autism-related expression pattern more closely resembled those of the
neurological conditions — Alzheimer’s and Parkinson’s — than the other psychiatric
ones. Neurological conditions, by definition, must have a
known physical signature in the brain, such as Parkinson’s characteristic loss
of dopaminergic neurons. Researchers have not found such a signature for
autism.
“This [similarity] just provides some kind of
additional direction we should look into,” Liu says. “Maybe one day we will
understand the pathology better.”
The findings were published in Molecular Psychiatry in November.
Two genes, CRH and TAC1, are the most commonly altered among the
conditions: CRH is downregulated in all of the conditions but Parkinson’s, and
TAC1 is downregulated in all but depression. Both genes affect the activation
of microglia, the brain’s immune cells.
Atypical microglial activation may be “derailing
normal neurogenesis and synaptogenesis,” Coe says, disrupting neuronal activity
similarly across the conditions.
Genes involved in astrocyte and synapse function are
similarly expressed in people with autism, schizophrenia or bipolar disorder, a 2018 study of postmortem brain tissue found. But microglial genes are
overexpressed in autism alone, that study found.
People with more
intensely upregulated immune genes could have a “neuroinflammatory condition,” says Michael Benros, professor and head
of research on biological and precision psychiatry at the University of
Copenhagen in Denmark, who was not involved in the work.
“It could be interesting to try to identify these
potential subgroups and of course provide them more specific treatment,” Benros
says.
Most of the
expression changes observed in the brain tissue samples did not appear in datasets
of gene expression patterns in blood samples from people with the same
conditions, the study shows. This “somewhat
surprising” finding indicates the importance of studying brain tissue,
says Cynthia Schumann, professor of
psychiatry and behavioral sciences at the University of California Davis MIND
Institute, who was not involved in the study.
“If you want to
know about the brain, you have to look at the brain itself,” Schumann says.
I am always reminding people not to think
that blood samples are going to tell them how to treat autism. The above commentary also highlights this
fact. If you want to know what is going
on in the brain, you have to look there or in spinal fluid. Looking just at blood samples may send an investigation
in completely the wrong direction. Spinal fluid flows around the brain and
spinal cord to help cushion them from injury and provide nutrients. Testing
spinal fluid requires an invasive procedure, parents do not like it and so it
is very rarely carried out until adulthood.
Time has then been lost.
Here is the link to the full paper and
some highlights I noted.
Neuroimmune transcriptome changes in patient brains of psychiatric and neurological disorders
Neuroinflammation has been implicated in multiple brain disorders but the extent and the magnitude of change in immune-related genes (IRGs) across distinct brain disorders has not been directly compared. In this study, 1275 IRGs were curated and their expression changes investigated in 2467 postmortem brains of controls and patients with six major brain disorders, including schizophrenia (SCZ), bipolar disorder (BD), autism spectrum disorder (ASD), major depressive disorder (MDD), Alzheimer’s disease (AD), and Parkinson’s disease (PD). There were 865 IRGs present across all microarray and RNA-seq datasets. More than 60% of the IRGs had significantly altered expression in at least one of the six disorders. The differentially expressed immune-related genes (dIRGs) shared across disorders were mainly related to innate immunity. Moreover, sex, tissue, and putative cell type were systematically evaluated for immune alterations in different neuropsychiatric disorders. Co-expression networks revealed that transcripts of the neuroimmune systems interacted with neuronal-systems, both of which contribute to the pathology of brain disorders. However, only a few genes with expression changes were also identified as containing risk variants in genome-wide association studies. The transcriptome alterations at gene and network levels may clarify the immune-related pathophysiology and help to better define neuropsychiatric and neurological disorders.
Multiple lines of evidence support the
notion that the immune system is involved in major “brain disorders,” including
psychiatric disorders such as
schizophrenia (SCZ), bipolar disorder (BD), and major depressive disorder
(MDD), brain development disorders such as autism spectrum disorder (ASD), and
neurodegenerative diseases such as Alzheimer's disease (AD), and Parkinson's
disease (PD). Patients with these brain diseases share deficits in cognition,
blunted mood, restricted sociability and abnormal behavior to various degrees.
Transcriptome studies have identified expression alterations of immune-related
genes (IRGs) in 49 postmortem brains of AD, PD, ASD, SCZ and BD separately.
Cross disorder transcriptomic studies further highlighted changes in IRGs. At
the protein level, several peripheral cytokines showed reproducible
disease-specific changes in a meta-analysis. Since brain dysfunction is
considered the major cause of these disorders, studying immune gene expression
changes in patient brains may reveal mechanistic connections between immune
system genes and brain dysfunction. Most previous studies were limited to the
analysis of individual disorders. There
is no comprehensive comparison of the pattern and extent of
inflammation-related changes in terms of immune constructs (subnetworks),
neuro-immune interaction, genetic contribution, and relationship between
diseases. Neuroinflammation, an immune response taking place within
the central nervous system, can be
activated by psychological stress, aging, infection, trauma, ischemia, and
toxins. It is regulated by sex, tissue type and genetics, many of which
are known disease risk factors for both psychiatric and neurological diseases. The primary function of
neuroinflammation is to maintain brain homeostasis through protection and
repair. Abnormal
neuroinflammation activation could lead to dysregulation of mood, social behaviors,
and cognitive abilities. Offspring who were fetuses when their mothers’ immune system was
activated (MIA) showed dopaminergic hyperfunction, cognitive impairment, and
behavioral abnormalities as adults. Alternatively, acute and chronic
neuroinflammation in adulthood can also alter cognition and behavior. In animal models, both adult and
developmental maternal immune activation in the periphery can lead to increases
in pro-inflammatory cytokines in the brain , similar to what is found in humans
with major mental illness.
Previous studies identified immune gene dysregulations in brains of
patients with several major brain disorders. For example, Gandal et al. found
that up-regulated genes and isoforms in SCZ, BD, and ASD were enriched in
pathways such as inflammatory response and response to cytokines. One brain
co-expression module up-regulated specifically in MDD was enriched for genes of
cytokine-cytokine interactions, and hormone activity pathways. The association
of neurological diseases such as AD and PD with IRGs has also been reported.
These studies examined the changes of immune system as a whole without going
into details of specific subnetworks, the disease signature, or genetic versus
environmental contribution. We hypothesize that expression changes of specific
subsets of IRGs constitute part of the transcriptome signatures that
distinguishes diseases. Since tissue specificity, sex and genetics all could
influence such transcriptome signatures, we analyzed their effects.
Furthermore, we expect that neurological diseases and psychiatric disorders
bear transcriptomic changes that may help to address how similar immunological
mechanisms lead to distinct brain disorders. The current boundary between
neurological diseases and psychiatric disorders is primarily the presence of
known pathology. Neurological diseases have more robust histological changes
while psychiatric disorders have more subtle subcellular changes. Nonetheless,
pathology evidence is always a subject to be revised with new research. To investigate immune-related signatures of
transcriptome dysregulation in brains of six neurological and psychiatric disorders,
we studied a selected list of 1,275 genes known to be associated with
neuroinflammation and interrogated their expression across disorders. We
collected and analyzed existing transcriptome data of 2,467 postmortem brain
samples from donors with AD, ASD, BD, MDD, PD, SCZ and healthy controls (CTL).
We identified the differentially expressed IRGs shared across disorders or
specific to each disorder, and their related coexpression modules (Fig. S1).
These genes and their networks and pathways provided important insight into how
immunity may contribute to the risk of these neurological and psychiatric
disorders, with a potential to refine disease classification.
The two most shared dIRGs are Corticotropin-releasing hormone (CRH) and Tachykinin Precursor 1 (TAC1), which were differentially expressed in five of the six diseases (Fig. 2D). They both involve innate immunity according to the databases we used and literature. CRH was downregulated in five of the six disorders; the exception was PD. CRH can regulate innate immune activation with neurotensin (NT), stimulating mast cells, endothelia, and microglia. TAC1 was down-regulated in five of the six disorders, the exception being MDD. TAC1 encodes four products of substance P, which can alter the immune functions of activated microglia and astrocytes. Independent RNA-seq data confirmed both CRH and TAC1 findings. These transcripts are also neuromodulators and have action on neurons so they have roles in addition to immune functions.
This indicated that even though immune dysfunction is widespread in the six disorders, signature patterns of the subset innate immune genes are sufficient to differentiate neurological from psychiatric disorders.
Disease-specific
IRMs in AD, ASD, and PD imply distinct biological processes.
We also searched for disease-specific IRMs for each disorder. We used rWGCNA to construct brain co-expression networks in the brains of each disorder and of controls, then compared them against each other to identify disease-specific IRMs (Fig.5A). Based on preservation results of one disease versus controls and against all other diseases (Fig. 5B, z-summary < 10), as well as immune gene enrichment results (Table S9; enrichment q.value < 0.05), we identified six disease-specific IRMs, including one for AD, three for ASD, and two for PD. We did not detect disease-specific IRMs for SCZ, BD, or MDD, which are considered psychiatric disorders. The disease-specific IRMs were enriched for various functions (Fig. 5C, Table S9). The AD specific IRM was enriched for neuron part (GO:0097458, q.value= 4.57E-4) and presynapse (GO:0098793, q.value = 4.57E-4). The PD-specific IRM was enriched for positive regulation of angiogenesis (GO:0045766, q.value = 9.65E-06) and secretory granule (GO:0030141, q.value= 220 6.31E-06). The ASD-specific IRMs were enriched for developmental biological processes such as negative regulation of cell proliferation and growth factor receptor binding.
Our reader Eszter will be pleased to see that the research links the differentially expressed genes more with Alzheimer’s than with Bipolar or Schizophrenia. She has noted the overlap in effective therapies between Alzheimer’s and autism.
We came up with four major findings of the neuroimmune system in brains of different neuropsychiatric disorders: 1) the innate immune system carries more alterations than the adaptive immune systems in the six disorders; 2) the altered immune systems interact with other biological pathways and networks contributing to the risk of disorders; 3) common SNPs have a limited contribution to immune-related disease risks, suggesting the environmental contribution may be substantial; and 4) the expression profiles of dIRGs, particularly that of innate immune genes, group neurodevelopment disorder ASD with neurological diseases (AD and PD) instead of with psychiatric disorders (BD, MDD, and SCZ) Dysregulation of the innate immune system is a common denominator for all six brain disorders. We found that more than half of the shared dIRGs and dIRG-enriched pathways were related to the innate immune system. The two most shared dIRGs, TAC1 and CRH, have known effects on innate immune activation(66, 67). Both genes were downregulated in patient brains. Additionally, TLR1/2 mediates microglial activity, which could contribute to neuronal death through the release of inflammatory mediators. Furthermore, innate immunity is critical in maintaining homeostasis in the brain. For example, the innate immune system has been reported to function in the CNS's resilience and in synaptic pruning throughout brain growth. When homeostasis is disrupted, the abnormal innate immunity may impact a wide range of brain functions.
Microglia are affected specifically in
autism and Alzheimer’s.
Microglia are highlighted in the immune changes in brains of AD and ASD in this study. Microglia is the major cell type participating in the brain’s immune system. Our analyses showed that the IRM12 coexpression module was enriched for microglia genes and associated with inflammatory transcriptional change in AD and ASD but not the other four diseases. Does this suggest that microglial dysfunction contributes more to AD and ASD than to the other disorders? The PsychENCODE study showed the microglial module upregulated in ASD and downregulated in SCZ and BD(16), but the fold changes in SCZ and BD were much smaller than that in ASD (Fig 7.B in original paper(16)). Larger sample size may be needed to detect microglia contribution to other disorders such as SCZ and BD.
Sex contributes to the disease-related immune changes too. Our results revealed sex-bias dysregulation of IRGs in brains of ASD and MDD but not in other disorders. These two disorders are known to have sex differences in prevalence. Previous studies also have suggested that sex differences in stress-related neuroinflammation might account for the overall sex bias in stress-linked psychiatric disorders, including female bias in MDD and male bias in ASD. We did not observe sex-biased IRGs in other diseases with known sex-biased prevalence, such as SCZ and AD suggesting that sex differences in SCZ and AD may not involve IRG changes.
Our results showed how immune system dysregulation may influence gene expression of the networked other non-immune genes and contribute to the pathology of these diseases specifically. Six disease-specific IRMs were detected in AD, ASD, and PD, showing that several functions of the immune-related networks also involved in corresponding disorders such as presynaptic related AD-IRM and Growth factor receptors-related ASD-IRMs. Presynaptic proteins are essential for synaptic function and are related to cognitive impairments in AD(85). Growth factor receptors and N-acetylcysteine are involved in the etiology of ASD. Secretogranin may be a pivotal component of the neuroendocrine pathway and play an essential role in neuronal communication and neurotransmitter release in PD (88). Furthermore, the immune system has been found to regulate presynaptic proteins(89), EGFR(90), and secretogranin(88). Our results indicate that alterations of the immune network can be disease-specific, affecting specific coexpression networks and driving distinct risk of each disorder.
To
our surprise, neurodevelopment disorder ASD was grouped with neurological
diseases (AD and PD) instead of with
psychiatric disorders (BD, MDD, and SCZ) according to the changes of IRGs,
particularly innate immune genes. Hierarchical clustering analysis based on the
effect size of IRGs placed the presumed psychiatric disorder ASD with other neurological
diseases. Previous studies
have reported that ASD patients exhibited more neurological and immunological
problems(99-102) compared to healthy people and to other brain disorders.
As more etiologies are uncovered, the traditional classification of these
diseases is increasingly challenged(93). Furthermore, we found that dIRGs
change more in neurological diseases (AD, PD, and ASD) than in the psychiatric
disorders (BD, SCZ, and MDD). It suggested that neuroimmunity dysregulation is
more severe in neurological diseases than in psychiatric disorders, led by AD.
Neuroimmunity may help to redefine disease classification in the future.
Conclusion
It is good to see there is excellent
research coming from China. Our reader Stephen has noted some interesting research underway in Russia. Look both East and West.
In today’s paper the focus was just on immune related genes. That in itself is a big step forward, since in this blog we are well aware of the key role of the immune system in autism.
In this study all of autism was
grouped together, when we know there will be many subgroups with totally different
profiles. In terms of treatment, you
would need to know which subgroup you are part of.
But it does tell you that part of your autism therapy is going to have to account for an altered immune status.
I would have to say that it does
follow Western research in getting a bit lost in the detail. We know that they found 275 of the immune
genes mis-expressed in autism.
How about presenting a simple list of
the 275 with whether the genes were over or under expressed ?
There are vast spreadsheets in the
supplemental data, but nothing as down to earth and common sense as that.
Instead the researchers were
preoccupied with overlaps between different conditions and churning out
statistics.
It is notable from the first paper I
mentioned today that one of the very top Chinese hospitals is actually trying
to apply personalized medicine using Rapamycin for autism and publishing a case
history. Bravo !!
A logical next step after trying to
modify mTOR would be to try epigenetic modification therapy using HDAC
inhibition.
One issue here is the age at which
therapy begins, not surprisingly some therapies need to commence at birth (or
ideally before) and do not give much effect later in life.
Romidepsin is one HDAC inhibitor used
in the research.
In the studies below Chinese
researchers in the US are making progress.
In 2018:
Autism's
social deficits are reversed by an anti-cancer drug
Using an epigenetic mechanism, romidepsin restored gene expression and alleviated social deficits in animal models of autism.
"In the autism model, HDAC2 is abnormally high,
which makes the chromatin in the nucleus very tight, preventing genetic
material from accessing the transcriptional machinery it needs to be
expressed," said Yan. "Once HDAC2 is upregulated, it diminishes genes
that should not be suppressed, and leads to behavioral changes, such as the
autism-like social deficits."
But the anti-cancer drug romidepsin, a highly potent
HDAC inhibitor, turned down the effects of HDAC2, allowing genes involved in
neuronal signaling to be expressed normally.
The rescue effect on gene expression was
widespread. When Yan and her co-authors conducted genome-wide screening at the
Genomics and Bioinformatics Core at UB's New York State Center of Excellence in
Bioinformatics and Life Sciences, they found that romidepsin restored the majority of the more than
200 genes that were suppressed in the autism animal model they used.
In 2021:
We found that combined administration of
the class I histone deacetylase inhibitor Romidepsin and the histone
demethylase LSD1 inhibitor GSK-LSD1 persistently ameliorated the autism-like social
preference deficits, while each
individual drug alone was largely ineffective.
We now
need some leading researchers/clinicians in China to actually translate this
approach to humans and see if it works.
Hopefully the PLA hospital in Beijing are keeping an eye out on what Zhen Yan
is up to at the University of Buffalo, NY.
With luck they will not wait 20 years to try it!